CycLex SIRT2 Deacetylase Fluorometric Assay Kit Ver.2
| Code | Size | Price |
|---|
| MBL-CY-1152V2 | 100 Assays | £541.00 |
Quantity:
Prices exclude any Taxes / VAT
Overview
Regulatory Status: RUO
Shipping:
Dry Ice
Storage:
-70°C
Images
Documents
Further Information
Background:
Sir2 is a conserved protein and was recently shown to regulate lifespan extension both in budding
yeast and nematode. In 2000, it was reported that the yeast Sir2 protein is a NAD(+)-dependent histone
deacetylase that plays a critical role in transcriptional silencing, genome stability and longevity. In
mammals, the homologs of Sir2 have been named sirtuins (SIRT), with seven members in a family
termed SIRT1 through SIRT7. They share a conserved central deacetylase domain but have different Nand
C termini and display distinct subcellular localization, suggesting different biological functions (1).
In contrast to SIRT1, mammalian SIRT2 is localized mainly in the cytoplasm. SIRT2 colocalizes with
the microtubule network and deacetylates Lys40 of alpha-tubulin (2). The same residue of alpha-tubulin
is also deacetylated by HDAC6, a class II HDAC, and deacetylation by HDAC6 leads to changes in
cellular motility (3).
A role for SIRT2 in cancer pathogenesis was demonstrated using a proteomic approach (4). The
SIRT2 gene, which is located at chromosome 19q13.2, lies within a region that is frequently deleted in
human gliomas, and levels of SIRT2 mRNA and protein expression are severely reduced in a large
fraction of human glioma cell lines (4). Ectopic expression of SIRT2 in these cell lines suppressed
colony formation and modified the microtubule network. These results indicate that SIRT2 may act as a
tumor suppressor and may function to control the cell cycle by acetylation of alpha-tubulin. It was
reported that SIRT2 inhibitor rescued alpha-synuclein toxicity and modified inclusion morphology in a
cellular model of Parkinson?s disease, however the exact mechanism remains uncertain.
However, the conventional method for measuring SIRT2 activity is very complicated and laborious. In
order to measure SIRT2 enzyme activity, it is necessary to prepare radioactive acetylated histone H4 as a
substrate. First, cells have to be labeled metabolically with radioactivity by adding radioactive acetic
acid to the culture medium. Second, radioactive acetylated histone has to be purified from the cells.
Following the reaction, it is necessary to extract and separate the radioactive acetyl group, which has
been released from acetylated histone, using ethyl acetate to measure the activity of the enzyme based on
the radioactivity.
Although a method for measuring the activity of deacetylase without the use of radioactive substances
was reported in recent years, owing to the use of fluorescent-labeled acetylated lysine as a substrate, the
reaction product must be separated from the intact substrate and the fluorescent intensity measured by
reverse phase HPLC. As mentioned above, these measurement systems are difficult to adapt for
processing many samples under a variety of conditions, because of their complicated operation. Thus a
simple system for biochemical analysis as well as for inhibitor screening without the use of radioactive
substances is preferred.
Description:
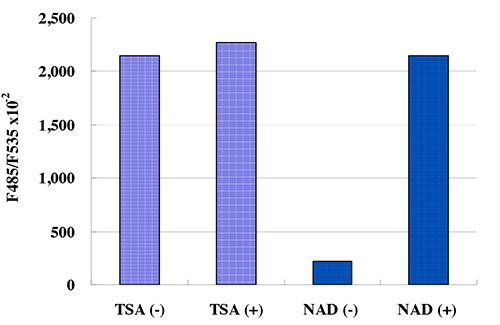
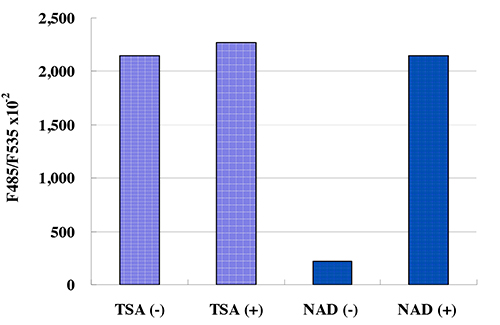
This kit is designed for the rapid and sensitive evaluation of SIRT2 inhibitors or activators using recombinant SIRT2 or purified SIRT2
Kit Components:
SIRT2 Assay Buffer, Fluoro-Substrate Peptide (0.2 mM), Fluoro-Deacetylated Peptide (0.2 mM), NAD (8 mM), Developer, Recombinant SIRT2, Stop Solution
Shelf Life:
1 year
Target:
SIRT2
References
1. North, B.J., and Verdin, E.; Sirtuins: SIRT2-related NAD-dependent protein deacetylases. Genome
Biol. 5: 224, 2004
2. North BJ, Marshall BL, Borra MT, Denu JM, Verdin E; The human SIRT2 ortholog, SIRT2, is an
NAD+-dependent tubulin deacetylase. Mol Cell 11:437-444, 2003
3. Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP;
HDAC6 is a microtubule-associated deacetylase. Nature 417: 455-458, 2002
4. Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic
CG, Kurimasa A, Oshimura M; Proteomics-based identification of differentially expressed genes in
human gliomas: down-regulation of SIRT2 gene. BBRC 309: 558-566, 2003
5. S Imai, CM Armstrong, M Kaeberlein, and L Guarente; Transcriptional silencing and longevity
protein Sir2 is an NAD-dependent histone deacetylase. Nature. 403: 795-800, 2000
6. Joseph Landry, Ann Sutton, Stefan T. Tafrov, Ryan C. Heller, John Stebbins, Lorraine Pillus, and Rolf
Sternglanz; The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases
Proc Natl Acad Sci U S A 97: 5807-5811, 2000
7. Jeffrey S. Smith, Carrie Baker Brachmann, Ivana Celic, Margaret A. Kenna, Shabazz Muhammad,
Vincent J. Starai, Jose L. Avalos, Jorge C. Escalante-Semerena, Charles Grubmeyer, Cynthia
Wolberger, and Jef D. Boeke; A phylogenetically conserved NAD+-dependent protein deacetylase
activity in the Sir2 protein family Proc Natl Acad Sci U S A 97, 6658-6663, 2000
8. H Vaziri, SK Dessain, E Ng Eaton, SI Imai, RA Frye, TK Pandita, L Guarente, and RA Weinberg;
hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 107: 149-159, 2001
9. J Luo, AY Nikolaev, S Imai, D Chen, F Su, A Shiloh, L Guarente, and W Gu; Negative control of p53
by Sir2alpha promotes cell survival under stress. Cell. 107: 137-148, 2001
10. E Langley, M Pearson, M Faretta, UM Bauer, RA Frye, S Minucci, PG Pelicci, and T Kouzarides;
Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21:
2383-2396, 2002
11. J Smith; Human Sir2 and the 'silencing' of p53 activity. Trends Cell Biol. 12: 404, 2002
12. CM Grozinger and SL Schreiber; Deacetylase enzymes: biological functions and the use of
small-molecule inhibitors. Chem Biol, 9: 3-16, 2002