CycLex SIRT3 Deacetylase Fluorometric Assay Kit Ver.2
| Code | Size | Price |
|---|
| MBL-CY-1153V2 | 100 Assays | £541.00 |
Quantity:
Prices exclude any Taxes / VAT
Overview
Regulatory Status: RUO
Shipping:
Dry Ice
Storage:
-70°C
Images
Documents
Further Information
Background:
Sir2 is a conserved protein and was recently shown to regulate lifespan extension both in budding
yeast and nematode. In 2000, it was reported that the yeast Sir2 protein is a NAD(+)-dependent histone
deacetylase that plays a critical role in transcriptional silencing, genome stability and longevity. There
are seven mammalian Sir2 homologs (1), all of which maintain the catalytic core domain of Sir2.
NAD-dependent deacetylase activity has been demonstrated for mammalian SIRT1, SIRT2, SIRT3,
SIRT5 and SIRT6 proteins.
The presence of NAD-dependent ADP-ribosylase and protein deacetylase activities of sirtuin proteins
suggests that they may function as sensors of metabolic or oxidative states of cells and regulate cellular
functions accordingly. Mammalian SIRT1, which resides in the nucleus, is the most closely related to
yeast Sir2. SIRT1 binds and deacetylates p53 (2, 3), NF-kappa B (4), forkhead transcription factors (5, 6),
and histones (7). SIRT1 also suppresses muscle differentiation in response to the redox state (8). SIRT2
is a cytoplasmic protein, which colocalizes with microtubules and deacetylates alpha-tubulin (9). SIRT2
abundance increases during mitosis, suggesting that the protein plays a role in cell cycle regulation (10).
Human SIRT3 is a mitochondria protein, with its N-terminal 25 amino acid residues responsible for its
mitochondrial localization (11, 12). Synthesized as an enzymatically inactive protein, human SIRT3 is
activated by mitochondrial matrix processing peptidase to active 28-kD active enzyme (13). These
observations suggest that the existence of a latent class III deacetylase that becomes catalytically
activated upon import into the human mitochondria.
However, the conventional method for measuring SIRT3 activity is very complicated and laborious. In
order to measure SIRT3 enzyme activity, it is necessary to prepare radioactive acetylated histone or p53
as a substrate. First, cells have to be labeled metabolically with radioactivity by adding radioactive acetic
acid to the culture medium. Second, radioactive acetylated histone has to be purified from the cells.
Following the reaction, it is necessary to extract and separate the radioactive acetyl group, which has
been released from acetylated histone, using ethyl acetate to measure the activity of the enzyme based on
the radioactivity.
Although a method for measuring the activity of deacetylase without the use of radioactive substances
was reported in recent years, owing to the use of fluorescent-labeled acetylated lysine as a substrate, the
reaction product must be separated from the intact substrate and the fluorescent intensity measured by
reverse phase HPLC. As mentioned above, these measurement systems are difficult to adapt for
processing many samples under a variety of conditions, because of their complicated operation. Thus a
simple system for biochemical analysis as well as for inhibitor screening without the use of radioactive
substances is preferred.
Description:
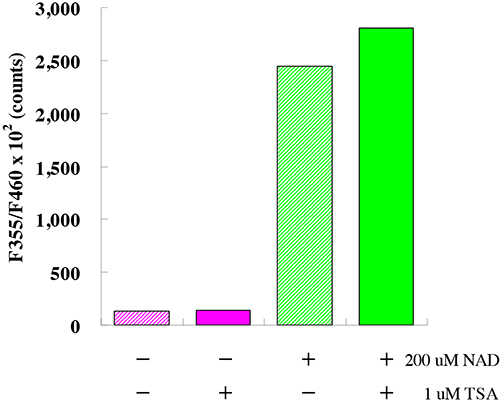
This kit is designed for the rapid and sensitive evaluation of SIRT3 inhibitors or activators using crude SIRT3 fraction or purified SIRT3.
Kit Components:
SIRT3 Assay Buffer, Fluoro-Substrate Peptide (0.2 mM), Fluoro-Deacetylated Peptide (0.2 mM), NAD (2 mM), Developer, Recombinant SIRT3, Stop Solution
Shelf Life:
1 year
Target:
SIRT3
References
1. Frye, R. A. Biochem. Biophys. Res. Commun. 260, 273?279, (1999)
2. Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. Cell 107,
137?148, (2001)
3. Langley, E., Pearson, M., Faretta, M., Bauer, U. M., Frye, R. A., Minucci, S., Pelicci, P. G., and
Kouzarides, T. EMBO J. 21, 2383?2396, (2002)
4. Yeung, F., Hoberg, J. E., Ramsey, C. S., Keller, M. D., Jones, D. R., Frye, R. A., and Mayo, M. W.
EMBO J. 23, 2369?2380, (2004)
5. Motta, M. C., Divecha, N., Lemieux, M., Kamel, C., Chen, D., Gu, W., Bultsma, Y., McBurney, M.,
and Guarente, L. Cell 116, 551?563, (2004)
6. Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., Tran, H., Ross, S. E.,
Mostoslavsky, R., Cohen, H. Y., Hu, L. S., Cheng, H. L., Jedrychowski, M. P., Gygi, S. P., Sinclair,
D. A., Alt, F. W., and Greenberg, M. E Science 303, 2011?2015, . (2004)
7. Vaquero, A., Scher, M., Lee, D., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. Mol. Cell 16,
93?105, (2004)
8. Fulco, M., Schiltz, R. L., Iezzi, S., King, M. T., Zhao, P., Kashiwaya, Y., Hoffman, E., Veech, R. L.,
and Sartorelli, V. Mol. Cell 12, 51?62, (2003)
9. North, B. J., Marshall, B. L., Borra, M. T., Denu, J. M., and Verdin, E Mol. Cell 11, 437?444, .
(2003)
10. Dryden, S. C., Nahhas, F. A., Nowak, J. E., Goustin, A. S., and Tainsky, M. A. Mol. Biol. Cell 23,
3173?3185, (2003)
11. Onyango, P., Celic, I., McCaffery, J. M., Boeke, J. D., and Feinberg, A. P. Proc. Natl. Acad. Sci. U. S.
A. 99, 13653?13658, (2002)
12. Schwer, B., North, B. J., Frye, R. A., Ott, M., and Verdin, E. J. Cell Biol. 158, 647?657, (2002)
13. Yang, Y. H., Chen, Y. H., Zhang, C. Y., Nimmakayalu, M. A., Ward, D. C., and Weissman, S.
Genomics 69, 355?369, (2000)