CycLex HDAC8 Deacetylase Fluorometric Assay Kit Ver.2
| Code | Size | Price |
|---|
| MBL-CY-1158V2 | 100 Assays | £562.00 |
Quantity:
Prices exclude any Taxes / VAT
Overview
Regulatory Status: RUO
Shipping:
Dry Ice
Storage:
-70°C
Images
Documents
Further Information
Background:
HDAC proteins are vital regulators of fundamental cellular events, including cell cycle progression,
differentiation, and tumorigenesis (1, 2). A small-molecule inhibitor of HDAC, trichostatin A (TSA),
arrests mammalian cells in both G1 and G2 (3, 4), while overexpression of HDAC1 in mouse cells
reduces their growth rate by lengthening the duration of G2 and M (5). TSA induces terminal
differentiation of mouse erythroleukemia cells and apoptosis of lymphoid and colorectal cancer cells. In
addition, TSA treatment of cells expressing the PML zinc finger protein derepresses transcription and
allows cells to differentiate normally (6). With this precedent, HDAC inhibitors are being actively
explored as potential agents for the treatment of certain forms of cancer (7-9).
The human HDACs are organized into three different classes based on their similarity to yeast HDAC
proteins (1, 2). Class I enzymes are ubiquitously expressed and include HDAC1, -2, -3, and -8, which
are homologous to the yeast RPD3 protein. Class II includes HDAC4, -5, -6, -7, -9, and -10, which are
similar to yeast HDA1 and are expressed in a tissue-specific manner. The Sir2-like class III HDACs,
including SIRT1 to -7, require NAD(+) for enzymatic activity.
It has been reported that HDAC8 is important for the growth of human tumor cell lines and has a
distinct inhibition pattern that differs from that of HDAC1 and -3, which both share 43% sequence
identity with HDAC8. These findings lead to open the way to the development of selective inhibitors of
this subtype as potential novel anticancer therapeutics.
However, the conventional method for measuring HDAC activity is very complicated and laborious.
In order to measure HDAC enzyme activity, it is necessary to prepare radioactive acetylated histone as a
substrate. First, cells have to be labeled metabolically with radioactivity by adding radioactive acetic
acid to the culture medium. Second, radioactive acetylated histone has to be purified from the cells.
Following the reaction, it is necessary to extract and separate the radioactive acetyl group, which has
been released from acetylated histone, using ethyl acetate to measure the activity of the enzyme based on
the radioactivity.
Although a method for measuring the activity of deacetylase without the use of radioactive substances
was reported in recent years, owing to the use of fluorescent-labeled acetylated lysine as a substrate, the
reaction product must be separated from the intact substrate and the fluorescent intensity measured by
reverse phase HPLC. As mentioned above, these measurement systems are difficult to adapt for
processing many samples under a variety of conditions, because of their complicated operation. Thus a
simple system for biochemical analysis as well as for inhibitor screening without the use of radioactive
substances is preferred.
Description:
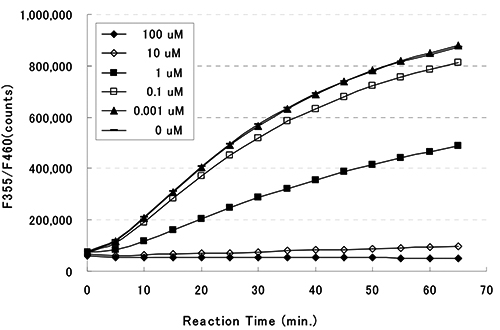
This kit is designed for the rapid and sensitive evaluation of HDAC inhibitors using recombinant HDAC8.
Kit Components:
HDAC Assay Buffer, Fluoro-Substrate Peptide (0.2 mM), Fluoro-Deacetylated Peptide (0.2 mM), Trichostatin A (200 uM), Developer, Recombinant HDAC8, Stop Solution
Shelf Life:
1 year
Target:
HDAC8
References
1. Thiagalingam, S., K. H. Cheng, H. J. Lee, N. Mineva, A. Thiagalingam, and J. F. Ponte. 2003. Histone
deacetylases: unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 983:84-100
2. Yang, X. J., and E. Seto. 2003. Collaborative spirit of histone deacetylases in regulating chromatin
structure and gene expression. Curr. Opin. Genet. Dev. 13:143-153
3. Ogryzko, V. V., T. H. Hirai, V. R. Russanova, D. A. Barbie, and B. H. Howard. 1996. Human
fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell
cycle dependent. Mol. Cell. Biol. 16:5210-5218.
4. Wharton, W., J. Savell, W. D. Cress, E. Seto, and W. J. Pledger. 2000. Inhibition of mitogenesis in
Balb/c-3T3 cells by trichostatin A. Multiple alterations in the induction and activation of
cyclin-cyclin-dependent kinase complexes. J. Biol. Chem. 275:33981-33987
5. Bartl, S., J. Taplick, G. Lagger, H. Khier, K. Kuchler, and C. Seiser. 1997. Identification of mouse
histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol. 17:5033-5043
6. He, L. Z., F. Guidez, C. Tribioli, D. Peruzzi, M. Ruthardt, A. Zelent, and P. P. Pandolfi. 1998. Distinct
interactions of PML-RAR{alpha} and PLZF-RAR{alpha} with co-repressors determine differential
responses to RA in APL. Nat. Genet. 18:126-135
7. Johnstone, R. W. 2002. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat.
Rev. Drug Discov. 1:287-299.[CrossRef][Medline]
8. Kelly, W. K., O. A. O'Connor, and P. A. Marks. 2002. Histone deacetylase inhibitors: from target to
clinical trials. Expert Opin. Investig. Drugs 11:1695-1713
9. Melnick, A., and J. D. Licht. 2002. Histone deacetylases as therapeutic targets in hematologic
malignancies. Curr. Opin. Hematol. 9:322-332
10. Gao, L., M. A. Cueto, F. Asselbergs, and P. Atadja. 2002. Cloning and functional characterization of
HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem.
277:25748-25755