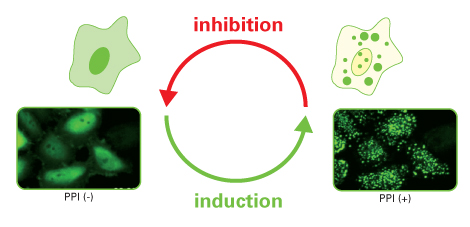
Fluoppi (Fluorescent-based technology detecting Protein-Protein Interaction) is a novel technology to detect Protein-Protein Interaction (PPI) in living cells with a high signal-to-noise ratio. The result is a bright, clearly analysable image. Fluoppi detects PPI as the absence or presence of fluorescent foci during inhibition or induction, respectively. The Fluoppi advantage is the ease of the construction of the PPI detection system with no need for optimising the linker.
Key Components
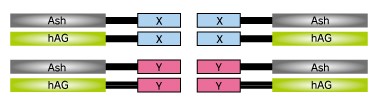
Fluoppi is a tag technology. Tetramer fluorescent protein (FP-tag) and Assembly helper tag (Ash-tag) are genetically fused to Protein X and Y, respectively. For the FP-tag, tetrameric fluorescent protein (e.g. Azami Green (hAG)) can be used. For the Ash-tag, PB1 domain of p62 can be used. PB1 is 102 amino acid length domain which has the capability of electrostatically homo-oligomerisation in a head to tail manner (Saio et al. 2010). In MBL’s in vitro Fluorescence correlation spectroscopy analysis, Ash-tag behaved as tetramer to octomer in diluted solution.
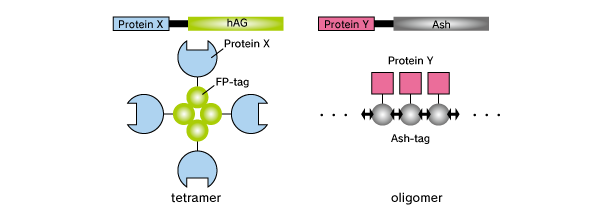
Product Format
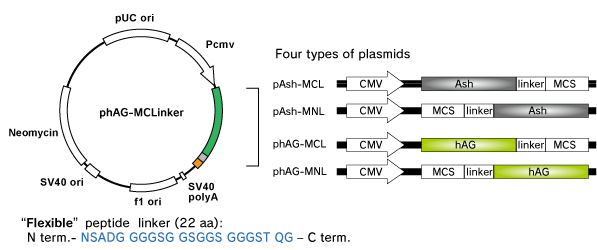
Fluoppi tags are able to work in both N and C terminal fusion. Several plasmid vectors include CMV promoter, Fluoppi tags, flexible peptide linker, Multiple Cloning Site (MCS) and Neomycin resistant gene.
Fluorescence Characteristics
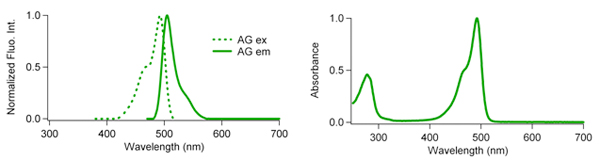
Azami Green Fluorescence Characteristics:
| Excit./Emiss.Maxima (nm) | Extinction coefficient (M-1cm-1) | Fluorescence quantum yield | pH sensitivity |
|---|---|---|---|
| 492/505 | 72,300 (492 nm) | 0.67 | pKa=5.0 |
Workflow
- Target gene amplification & restriction enzyme cut. At first, proteins X & Y of your interest are fused to FP-tag and Ash-tag respectively. MBL recommends to prepare all the eight possible constructs to identify the best workable combination.
- Ligation & sequencing
- Transfection. Co-transfect the pairs of plasmids.
- Imaging. Because fluorescent signal of Fluoppi is very high, conventional fluorescence microscopy can be used to image the cell. If the proteins interact with each other upon expression, fluorescent Puncta will be detected. Formation of Puncta is reversible so that they can be dissociated and the fluorescent signal will spread over the cell by PPI inhibitors, and vice versa by PPI inducer.
Properties
Localisation
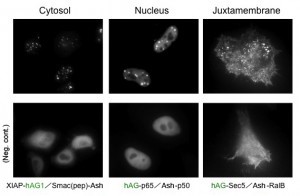
Because location of Puncta is not restricted to a specific site inside the cell, Fluoppi can detect PPIs at several subcellular localisations such as cytosol, nucleus, and juxtamembrane. The upper pictures represent Puncta at several subcellular localisations, and the lower pictures are negative controls which express hAG tagged protein and Ash-tag without fusing any proteins. The images of juxtamembrane are taken by Total Internal Reflection Fluorescence Microscopy (TIRFM).
PPI modulator (Induction)
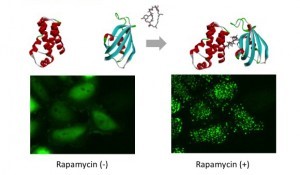
mTOR (FRB domain) and FKBP12 is a well known PPI whose interaction is induced by Rapamycin. The pair of, mTOR(FRB)-hAG and Ash-FKBP12 was selected from eight possible combinations in a viewpoint of foci formation efficiency. After adding Rapamycin small Puncta were gradually formed in several minutes.
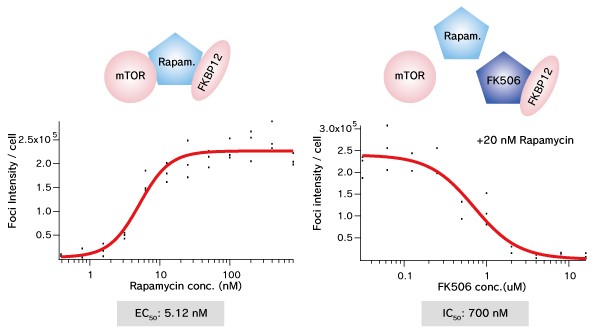
Dose dependent Puncta formations of Rapamycin (left) or competitive inhibition of Rapamycin by FK506 (right) were analysed. Cells were treated with Rapamycin for 60 min in 37°C CO2 incubator followed by PFA fixation. Images were captured by ORCA-ER(Hamamatsu photonics) on IX71 fluorescence microscopy (Olympus). The spot detector plugin from ICY platform (Institut Pasteur) and IGOR software (HULINKS Inc.) were used for this analysis.
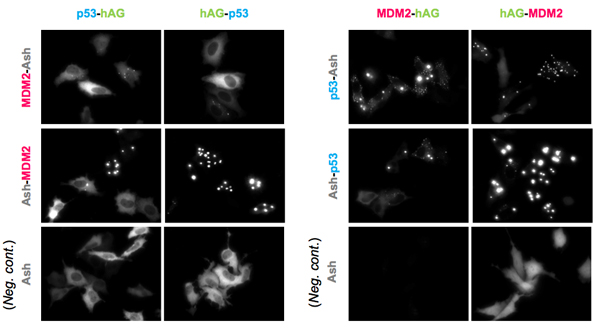
PPI modulator (Inhibition)
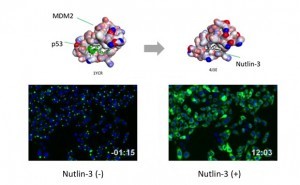
p53-MDM2 is a famous target of PPI inhibitors in the field of anti cancer drug development. MBL applied this PPI to Fluoppi for demonstration. First, hAG-MDM2 and Ash-p53 was selected from the 8 pairs as represented above, then stable CHO-K1 cell line was established by using two selection marker; G418 and Hygromycin. Fluoppi plasmids including Hygromycin resistance gene is under released at present.
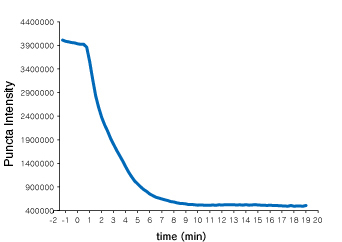
Time course of Puncta dissociation after adding Nutlin-3 was analysed. Y axis represents the fluorescent intensity of foci.
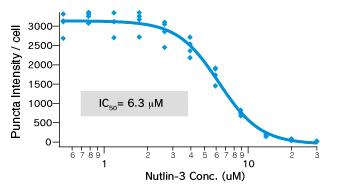
Dose dependent Puncta dissociations of Nutlin-3 were analysed. Cells were treated with Nutlin-3 for 30 min followed by PFA fixation. Images were measured by InCellAnalyzer1000(GE healthcare).
Originally posted on: https://www.mblbio.com/bio/g/product/flprotein/fluoppi.html#fluoppi
Caltag Medsystems is the distributor of MBL products in the UK and Ireland. If you have any questions about these products, please contact us.
