Introduction
In the field of molecular biology, microRNAs (miRNAs) have emerged as pivotal players in the intricate symphony of gene regulation. These small RNA molecules, typically composed of 20 to 25 nucleotides, play a critical role in post-transcriptional gene silencing. Understanding their spatiotemporal expression patterns within cells and tissues is paramount for unravelling their functional significance. This is where miRNA in situ hybridisation comes into play.
miRNA Nomenclature
| miRNA Names | Notes on Nomenclature |
|---|---|
| hsa-miR-XX vs. mmu-miR-XX vs. rno-miR-XX | The first three letters indicate the organism the miRNA is found in. hsa = human, mmu = mouse, rno = rat. |
| hsa-mir-XX vs. hsa-miR-XX | Capitalisation indicates whether it is the mature or precursor miRNA. mir = precursor sequence, miR = mature sequence. |
| hsa-miR-XX-5p vs. hsa-miR-XX-3p | The guide and passenger miRNAs are processed from the pre-miRNA. The guide miRNA is found in higher abundance than the passenger miRNA. Older alternative to the -5p and -3p nomenclature. |
| hsa-miR-XX-1 vs. hsa-miR-XX-2 vs. hsa-miR-XX | Mature miRNA sequences that are identical, but are originally transcribed from different genes and have distinct precursor sequences. As the sequences are identical, they may be referred to without the numerical suffix. |
| hsa-mir-XX-1 vs. hsa-mir-XX-2 | Precursor miRNAs that are different, but are processed into an identical miRNA. |
| hsa-miR-XXa vs. hsa-miR-XXb | Closely related mature miRNAs (differ by only one or two nucleotides). |
Bernardo, et al., (2012)
What is miRNA In Situ Hybridisation?
miRNA in situ hybridisation is a powerful technique that allows researchers to visualise and localise miRNAs within cells and tissues. It is based on the principle of hybridisation, where a complementary probe specifically binds to the target miRNA, enabling its detection and spatial localisation.
Advancements in In Situ Hybridization (ISH) Technology
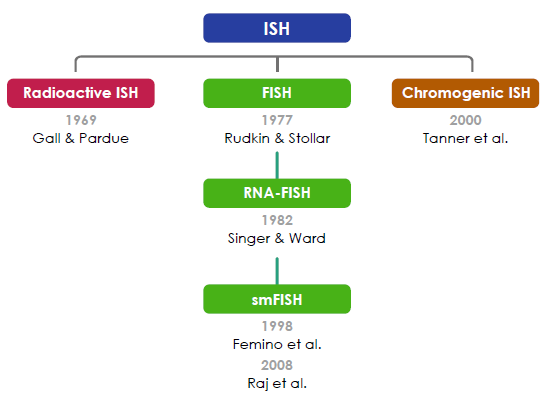
In situ hybridisation (ISH) was first performed by Gall & Pardue (1969) using radioactive probes. Fluorescent ISH (FISH) against DNA was first performed by Rudkin & Stollar (1977). FISH against RNA(RNA-FISH) was first performed by Singer & Ward (1982). RNA-FISH that could be used to resolve individual mRNA transcripts was first performed by Femino et al. (1998) and later improved upon in whole mount tissue by Raj et al. (2008). Horseradish peroxidase-based chromogenic (or colourimetric) ISH was later introduced by Tanner et al. (2000) as an alternative FISH without the need for a fluorescence microscope.
Young et al., (2020).
How Does It Work?
The process begins with the design of miRNA-specific probes. These probes are typically short RNA sequences complementary to the miRNA of interest. Importantly, they are labelled with detectable markers such as fluorescent dyes or digoxigenin tags for visualisation.
Tissue or cell samples are first fixed and processed, followed by the application of labelled probes. The probes hybridise with the target miRNA, forming a stable double-stranded structure. Any unbound probes are washed away. The location of the hybridisation signal indicates the expression site of the miRNA in the sample.
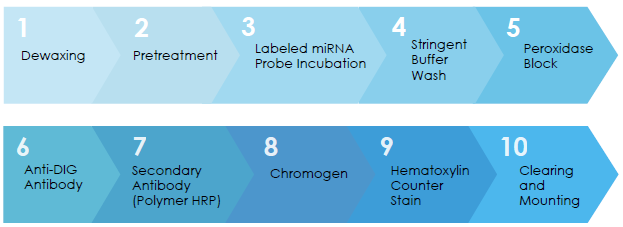
Workflow for miRNA Chromogenic In Situ Hybridisation in FFPE Tissue Sections
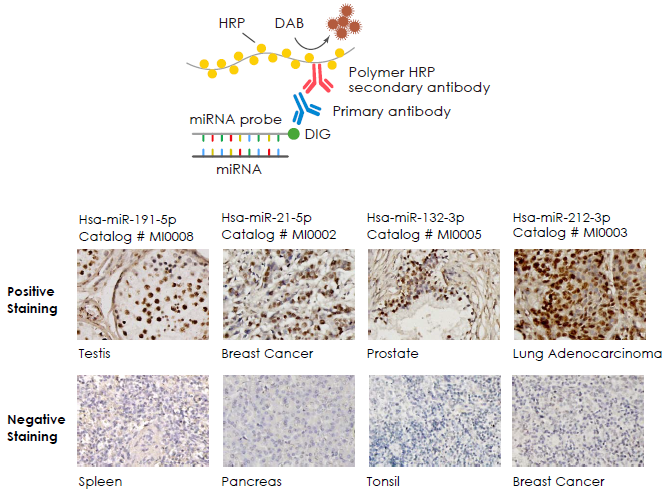
Examples
Applications of miRNA In Situ Hybridisation
Basic Research Applications
- Spatial Expression Patterns: miRNA in situ hybridisation allows researchers to determine the spatial expression patterns of specific miRNAs within tissues or cells. This spatial information is crucial for understanding the roles of miRNAs in development, tissue homeostasis, and disease.
- Cellular Co-localisation: Researchers can use miRNA in situ hybridisation in conjunction with immunohistochemistry to examine the co-localisation of miRNAs with specific cell markers, revealing their roles in cell-type-specific functions.
- Disease Research: Investigating miRNA expression patterns in disease tissues provides insights into the involvement of miRNAs in pathogenesis. Altered miRNA expression is often associated with various diseases, including cancer and neurodegenerative disorders.
- Developmental Biology: miRNA in situ hybridisation is indispensable in developmental biology, aiding in unravelling the temporal and spatial regulation of miRNAs during embryonic development and organogenesis.
Clinical Applications Potential
- Cancer Diagnosis and Prognosis: miRNA probe in situ hybridisation can be used to analyse miRNA expression in tumour tissues, aiding in the diagnosis of different types of cancer and the assessment of patient prognosis.
- Cancer Therapy: Understanding miRNA expression patterns in tumours can provide the basis for miRNA gene therapy, including the use of miRNA mimics or inhibitors to regulate miRNA functions.
- Neurological Diseases: Research on the role of miRNAs in neurodegenerative diseases may lead to the development of new neuroprotective treatment approaches.
- Cardiovascular Diseases: miRNA probe in situ hybridisation can be employed to study miRNA expression and regulation in cardiovascular diseases, identifying potential therapeutic targets.
- Metabolic Diseases: Investigating miRNA involvement in metabolic diseases can help understand the mechanisms of diseases such as obesity, diabetes, and lipid metabolism disorders and develop related treatment strategies.
Sempere et al., (2020)
Ho et al., (2022)
Originally posted by Abnova on https://www.abnova.com/en-global/newsletter/newsletter/content/newsletter2023-11-01
Caltag Medsystems is the distributor of Abnova products in the UK and Ireland. If you have any questions about these products, please contact us.
